
Title: request
emailaddress:
pid: 64843784
Status: finished
NB: The data will be kept for two weeks, after which it will be deleted without further notice.
RAPHAEL output
PDB: 1hm9A Ratio Alpha helix: 0.218340611353712 Ratio Beta strand: 0.299126637554585 Ratio Coils: 0.482532751091703 Total score: 1.4993 Variance in period matrix: 7.50864 Variance in sequence seperation for 6 Å contacts: 9.76331000000002 Predicted repeat length: 25.4246 Minimum distance between N and C terminus divided by protein length: 0.0950245000000004 Concentration of 15 Å contacts with sequence seperation > 55 : 0.172785999999999 Structure status: REPEAT SVM SCORE: 10.406811...... (-1≥score≥1 indicates prediction accurate and precise) |
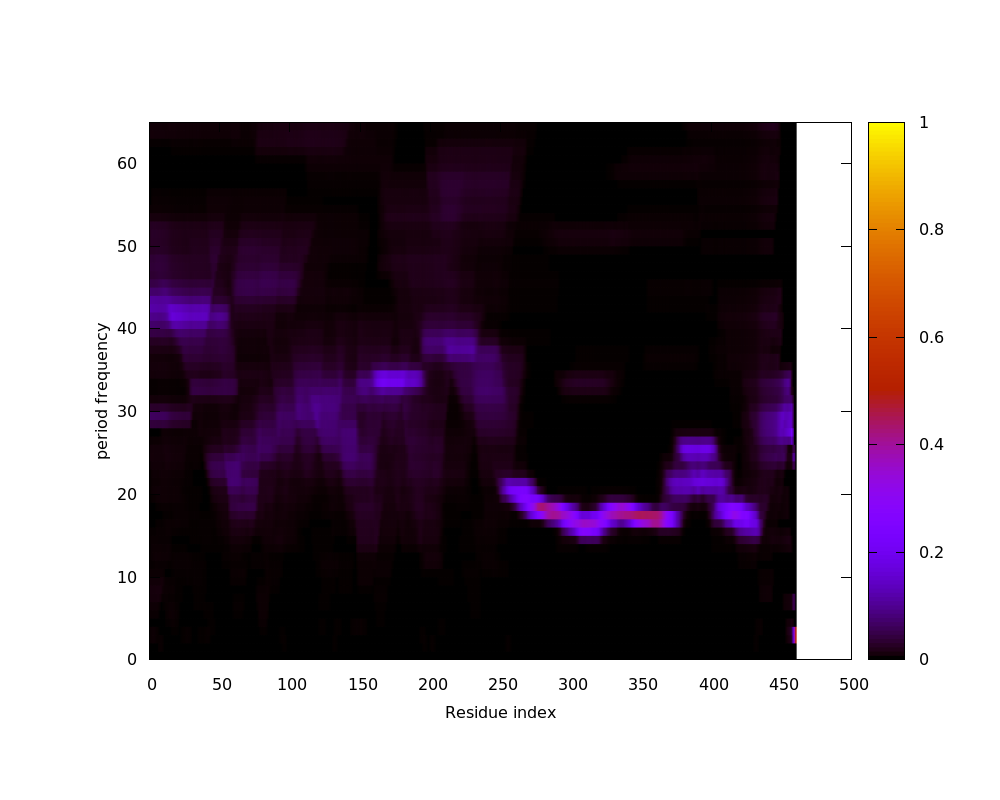 |
Sequence info: | 10 20 30 40 50 60 70
SNFAIILAAGKGTRMKSDLPKVLHKVAGISMLEHVFRSVGAIQPEKTVTVVGHKAELVEEVLAGQTEFVT
.EEEEEE.....GGG..SS.GGGSEETTEEHHHHHHHHHHTT..SEEEEEE.TTHHHHHHSSSSSSEEEE
0000011100000000000000000000000111111000000000111111111110000011001111
80 90 100 110 120 130 140
QSEQLGTGHAVMMTEPILEGLSGHTLVIAGDTPLITGESLKNLIDFHINHKNVATILTAETDNPFGYGRI
.SS...HHHHHHTTHHHHTT..SEEEEEETT.TT..HHHHHHHHHHHHHTT.SEEEEEEE.S..TTS.EE
1111110000011111000000000111100000000000000011000000000000000001111111
150 160 170 180 190 200 210
VRNDNAEVLRIVEQKDATDFEKQIKEINTGTYVFDNERLFEALKNINTNNAQGEYYITDVIGIFRETGEK
EE.TT..EEEEE.TTT..TTGGG..EEEEEEEEEEHHHHHHHHTT..S.STT.S..TTHHHHHHHHHT..
1111000000000111111100011111111111000000000001111100011000000111001100
220 230 240 250 260 270 280
VGAYTLKDFDESLGVNDRVALATAESVMRRRINHKHMVNGVSFVNPEATYIDIDVEIAPEVQIEANVILK
EEEEE.SSGGGG....SHHHHHHHHHHHHHHHHHHHHHTT.EES.GGG.EE.TT.EE.TT.EE.SS.EEE
0000000000000110000000000000011111111000001111111111111111111111111111
290 300 310 320 330 340 350
GQTKIGAETVLTNGTYVVDSTIGAGAVITNSMIEESSVADGVTVGPYAHIRPNSSLGAQVHIGNFVEVKG
SS.EE.TT.EE.TT.EEES.EE.TT.EE.S.EEES.EE.TT.EE.SS.EE.SS.EE.TT.EEEEEEEEES
1111111111111111111111111111111111111111111111111111111111111111111111
360 370 380 390 400 410 420
SSIGENTKAGHLTYIGNCEVGSNVNFGAGTITVNYDGKNKYKTVIGDNVFVGSNSTIIAPVELGDNSLVG
.EE.TT.EEEEEEEEESEEE.TT.EE.TT.EEE.B.SS.B..EEE.TT.EE.TT.EEESS.EE.TT.EE.
1111111111111111111111111111111111111110000111111111111111111111111111
430 440 450 458
AGSTITKDVPADAIAIGRGRQINKDEYATRLPHHPKNQ
TT.EE.S.B.TT.EE..S......TTGGGGSTTSGGG.
11111111111111111111110000000000000000
|
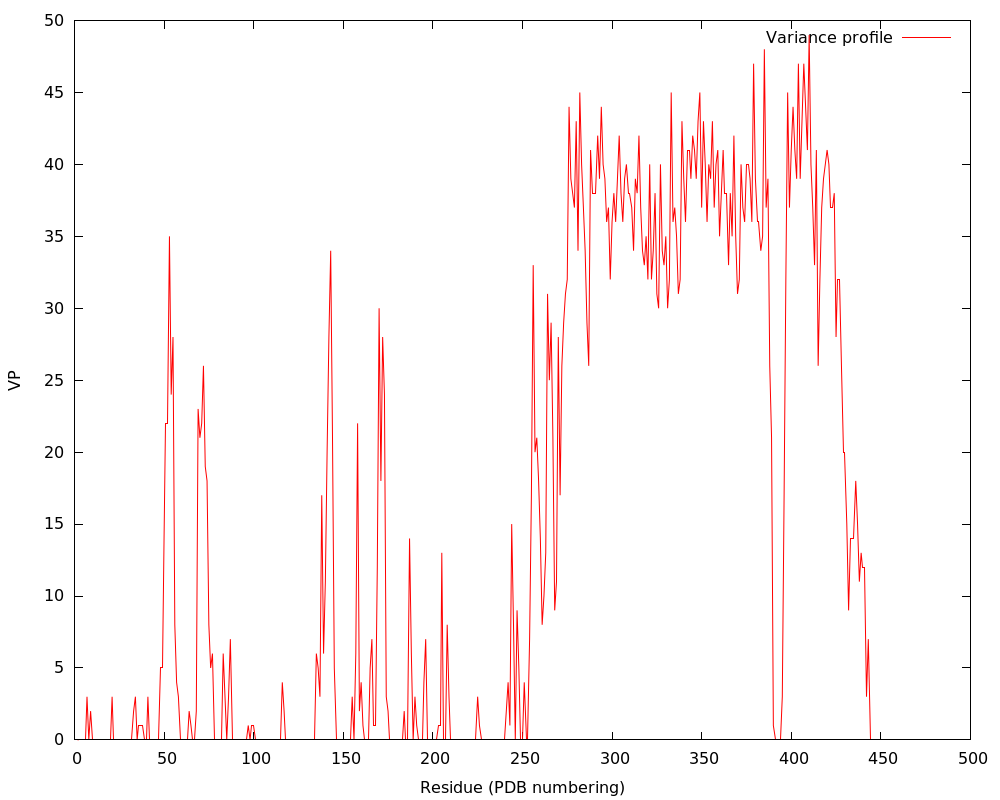 |